Prioritizing Compounds, Step 3: Predicting the Mutagenicity of the Selected Compounds
Predicting the Mutagenicity of the Selected Compounds
To add a further criterion to be used when selecting our drug candidates, we predict the compounds’ mutagenicities. To do so, we’ll use the ToxTree Benigni/Bossa rules for mutagenicity and carcinogenicity (Benigni et al., Mechanistic QSAR of aromatic amines: new models for discriminating between mutagens and nonmutagens, and validation of models for carcinogens, Environ Mol. Mutag. 48:754-771 (2007).). The URL of this model is http://apps.ideaconsult.net:8080/ambit2/model/8.
Analogously as you have done for the Cramer rules, navigate to the SuperService page in Ambit (http://apps.ideaconsult.net:8080/ambit2/algorithm/superservice), enter the dataset URI of the TCAMS dataset with the "?max=15000" appendix (http://apps.ideaconsult.net:8080/ambit2/dataset/584486?max=15000) as well as the URI of the ToxTree Benigni/Bossa model (http://apps.ideaconsult.net:8080/ambit2/model/8) to the appropriate fields and click "Run".
It is also possible to restrict calculations to the filtered list of the previous step only. In this case, use http://apps.ideaconsult.net:8080/ambit2/dataset/584486?property=http://apps.ideaconsult.net:8080/ambit2/feature/22254&search=Low+(Class+I) as the dataset URI.
OpenTox models store the prediction results (prediction features) again under data columns with a unique URI. These are available via http://host/model/{id}/predicted , which in our example corresponds to http://apps.ideaconsult.net:8080/ambit2/model/8/predicted.
For our purpose, we select the columns “Structural Alert for genotoxic carcinogenicity“ (http://apps.ideaconsult.net:8080/ambit2/feature/21858) and ”Structural Alert for nongenotoxic carcinogenicity“ (http://apps.ideaconsult.net:8080/ambit2/feature/21859). As before, we add data columns for these structural alerts to our Cramer-class filtered list of compounds (use the URI from the previous step as a starting point), again using the feature_uris[] method. The resulting URL is:
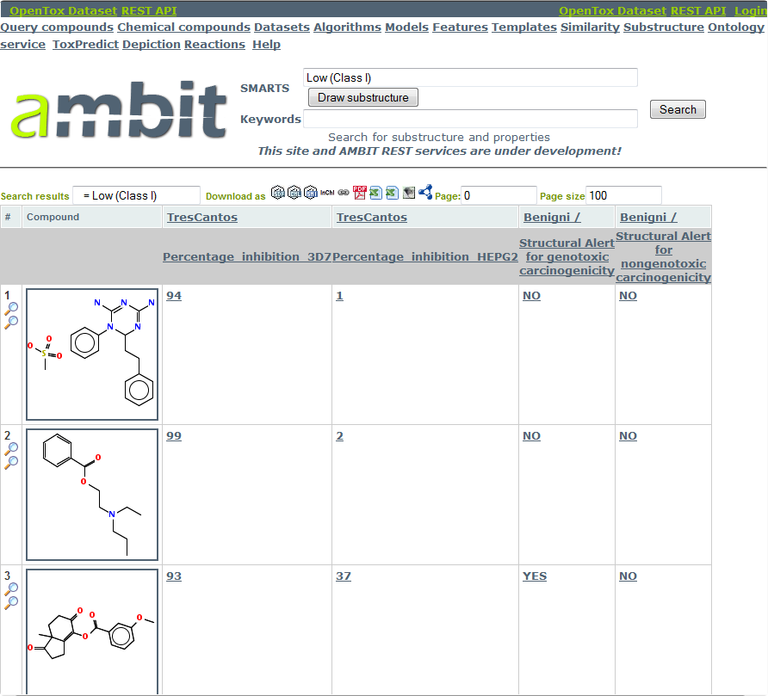
In the following examples, we’ll consider the second compound in the image below as our antimalarial drug candidate. It is a Cramer class I compound that inhibits growth of P. falciparum 3D7 by 99% at the concentration tested (2µM), has a very low human cytotoxicity and no structural alerts for carcinogenicity. (You may choose a different compound).
Similarly to datasets and models, each compound in OpenTox services also has its unique URI. You can find the URI of a compound by clicking on its 2D structure, and stripping off the “?media=text/html” part at the end of the URI in the address bar of the browser. Alternatively, you can right-click the 2D drawing, select "Copy Link Location" (Firefox) or "Copy Shortcut" (Internet Explorer). When pasting the saved URI, again strip the “?media=text/html” part at the end of the URI.
The URL of the compound selected above is http://apps.ideaconsult.net:8080/ambit2/compound/212976/conformer/621832.