Apply Bioclipse and OpenTox to modify Chemical Structure to remove Toxicity
This tutorial features a hands-on session with Bioclipse and the Decision Support feature together with the recently developed OpenTox plugin. Users will modify chemical structures with the aim to overcome chemical liabilities.
Download and install Bioclipse
Download and install Bioclipse with the pre-packaged OpenTox and Decision Support plugins from http://www.bioclipse.net/opentox and make sure you download one of the versions marked with “Packages containing also the Opentox Decision-Support plugin”.
Extract the downloaded ZIP-file to any place on your computer. To start Bioclipse (make sure you are connected to the internet), navigate inside the extracted folder and find the executable file for your operating system.
Getting Started
On first startup, click the menu “Window > Open Perspective > Other…” then select "Decision Support" from the dialog. This opens up the Decision Support view of Bioclipse displaying 3 endpoints with local models (running on the local computer) and several models in the OpenTox endpoint, running on the OpenTox servers in Europe.
Some of the OpenTox models are only available to registered users of OpenTox. Therefore, enter your OpenTox credentials in Bioclipse (if you don't have an OpenTox account yet, get one at http://opentox.org/join_form). To set up your OpenTox account in Bioclipse, go through the following steps:
- "File > Log in". A new window will open.
- In the new window that appears, select "Create new Keyring user...". A new window will open.
- In the new window, type a username and password, and click "OK". A new window will open.
- In the new window, click the "Add account" button at the bottom-left. A new window will open.
- In the new window, give the account a name (arbitrary), and make sure the Account type is "OpenTox OpenSSO Account". Click OK.
- This brings you back to the "Keyring User Properties" window, with the just created account highlighted.
- Now you need to enter your OpenTox username and password.
- First, click on "username" in the "Property" column. Then click on the empty field under "Value". Now type in your OpenTox user name.
- Second, click on "password" in the "Property" column. Then click on the empty field under "Value". Now type in your OpenTox password.
- Click "OK".
Importing Compounds
All files in Bioclipse must lie within a project; you can create one by using menu “File > New..” and then in the dialog selecting “Project” under the “General” tab. Now, drag and drop files (mol, cml, sdf) into the project. Double-click on a chemical structure to open it for editing. Note how the models become active in the Decision Support View.
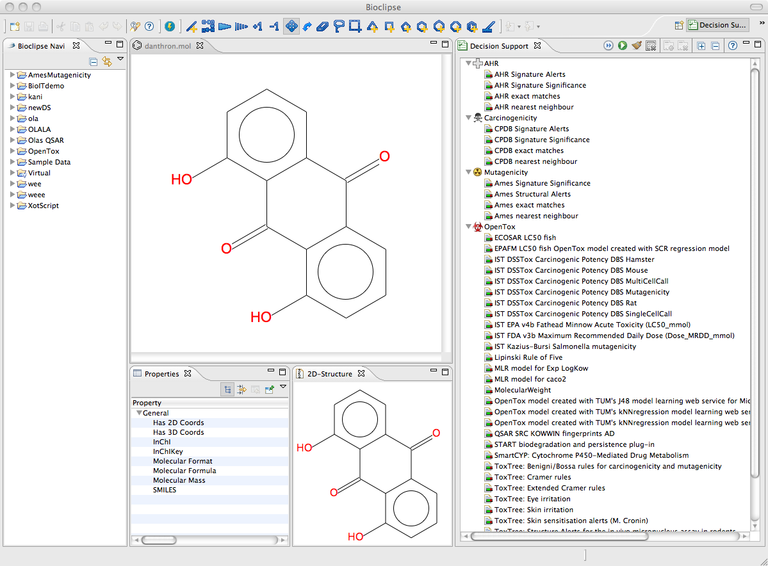
The toolbar in the Decision support view to the right has a play button (RUN). Click it to run all predictive models for the compound.
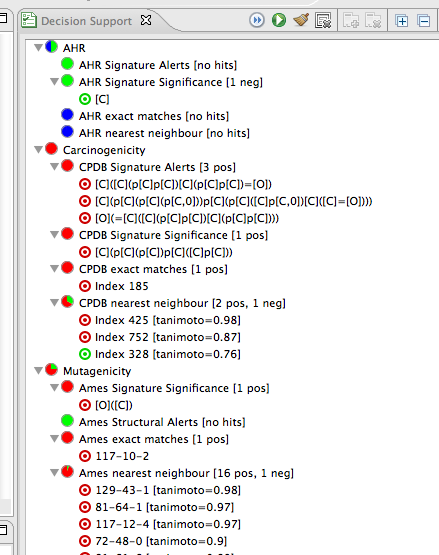
These are the results for the different models in the predictions. Click on one of the lines with a bullseye icon (in the figure above e.g. "[O]([C])" under "Ames Signature Significance [1 pos] within the Mutagenicity model. This will highlight the atoms in your molecule responsible for this prediction.
You can make changes to the compound and try again. Scroll down to see the OpenTox predictions.
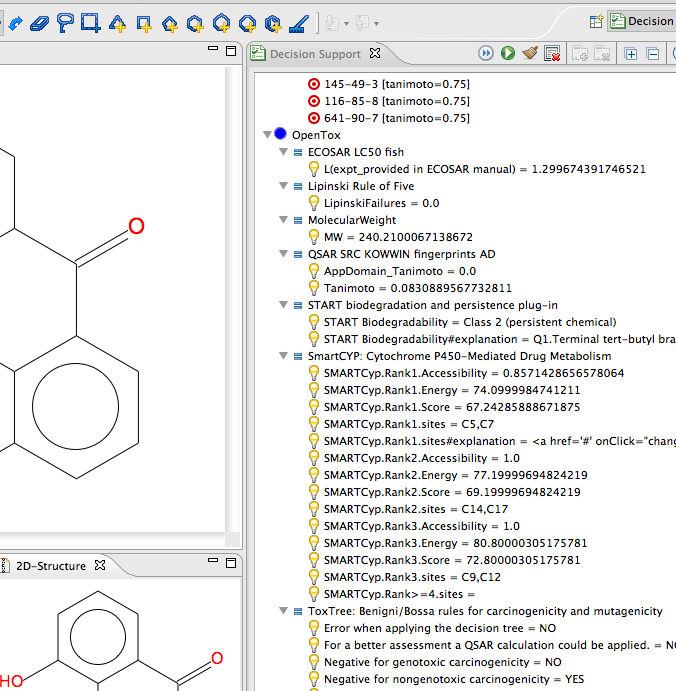
View it in Action
This short Screencast illustrates the Decision Support functionality in action:
http://opentox.org/data/documents/development/tutorialfiles/downloads/CalculateAsYouDraw-OT.mov/view
Additional information
In May 2011 we held an online tutorial session. You can download a recording of the tutorial at
http://opentox.org/data/documents/development/tutorialfiles/downloads/2011-05-25_Bioclipse-OpenTox-Integration.wmv
Scripting
If you are interested in learning how to use and develop scripts in Bioclipse to access OpenTox services, visit the following page:
http://chem-bla-ics.blogspot.com/2011/05/my-slides-for-this-afternoons-bioclipse.html
Evaluation Form
Complete the evaluation form following this link: Evaluation Form

