Prioritizing Compounds, Step 4: Predicting Sites of Cytochrome P450 Metabolism
Predicting Sites of Cytochrome P450 Metabolism
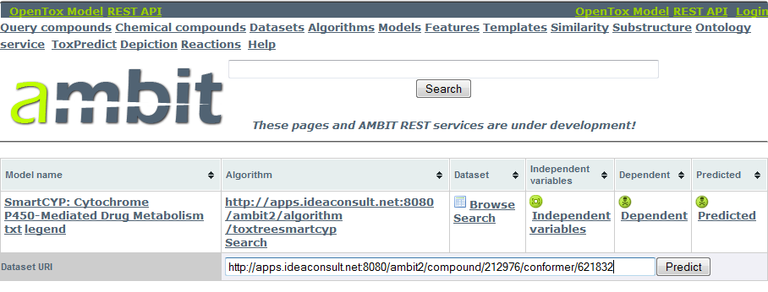
The URI of our drug candidate (http://apps.ideaconsult.net:8080/ambit2/compound/212976/conformer/621832) will be used to submit this compound to a model predicting cytochrome P450 sites of metabolism, namely
- SmartCYP (http://apps.ideaconsult.net:8080/ambit2/model/48, Rydberg P. et al., SMARTCyp: A 2D Method for Prediction of Cytochrome P450-Mediated Drug Metabolism, ACS Medicinal Chemistry Letters 2010, 1(3), 96-100) and
Model prediction is done by navigating to the model's URI and entering the compound URI into the "Dataset URI" field, followed by clicking the "Predict" button.
Clicking "Predict" leads to the task page of the model calculation. Click on the link under "Name" to refresh the status of your task:
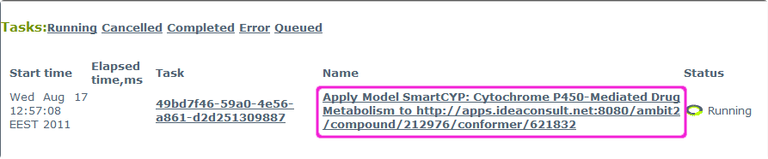
Repeat this until the status changes to "Completed":
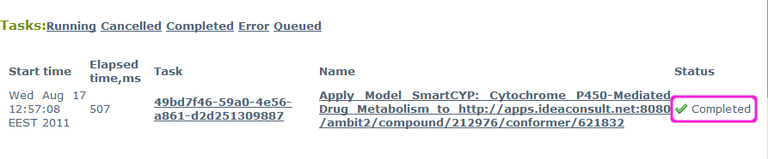
When completed, click again on the link under Name, to view the results. They are also available directly at this link:
http://apps.ideaconsult.net:8080/ambit2/compound/212976/conformer/621832?feature_uris[]=http://apps.ideaconsult.net:8080/ambit2/model/48/predicted
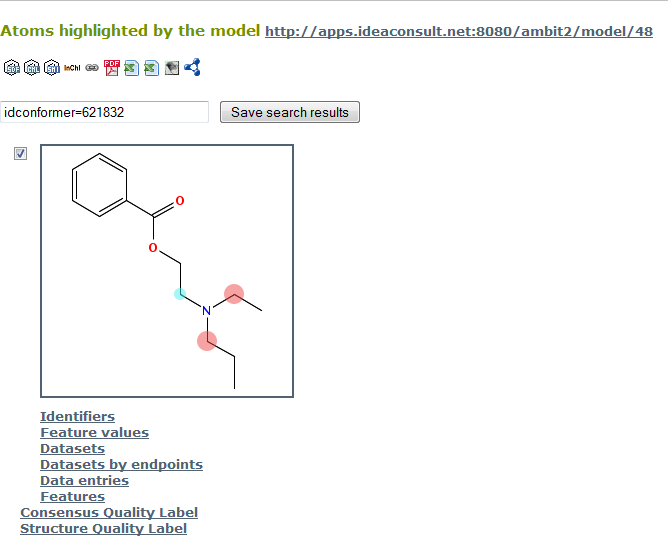
The results consist of information on which atoms are of rank 1, 2, 3 or lower. Higher rank means a more labile site. This information will be best viewed graphically, which could be achieved by the following URL
http://apps.ideaconsult.net:8080/ambit2/compound/212976/conformer/621832?model_uri=http://apps.ideaconsult.net:8080/ambit2/model/48 (see the following Figure).
The colour code for the result can be found by clicking on the “legend” link on the model page (http://apps.ideaconsult.net:8080/ambit2/model/48):
The "legend" link will display the following colour code:
Back to the Drug Discovery Predictive Toxicology Tutorial Overview
Back to the Tutorials page