Prioritizing Compounds, Step 1: Predicting Oral Toxicity
Predicting Oral Toxicity
Important to understand the OpenTox framework is that everything is referred to by its URI. Every molecule, every feature, every algorithm, every model, etc., everything is identified by a URI. A key step, therefore, for a calculation carried out in OpenTox is to find out the URIs of the necessary "ingredients". In the case of applying a model to a dataset to obtain predictions, the URIs to identify are the dataset URI and the model URI.
In this tutorial we make use of the Ambit2 implementation of the OpenTox framework (http://apps.ideaconsult.net:8080/ambit2).
Find the TCAMS antimalarial dataset at http://apps.ideaconsult.net:8080/ambit2/dataset?search=^T. This page lists all datasets whose names begin with the letter "T". Find the dataset called "TCAMS Malaria Box". We will first predict oral toxicity for this dataset. Start by clicking on the TCAMS Malaria Box dataset link. The URL in the browser should read http://apps.ideaconsult.net:8080/ambit2/dataset/584486?page=0&pagesize=10. You can browse the compounds. Note that only 10 compounds are displayed. The dataset contains well over 13000 compounds, however. If you observe the address in your browser closely, you will notice the "?page=0&pagesize=10" part. This means that we want to display 10 compounds per page, and want to have the first page (page 0) displayed. To see the next 10 compounds, you can change page=0 to page=1 in the address bar, or simply use the appropriate field at the top of the dataset to change from page 0 to page 1. Similarly, change the pagesize to e.g. 100 if you want to have 100 compounds displayed per page.
From the address bar of your browser, you can also identify now the URI of the dataset. It is whatever comes before the "?" character, in this case http://apps.ideaconsult.net:8080/ambit2/dataset/584486. Note the the URI including the "?page=0&pagesize=100" part is also a valid dataset URI in OpenTox. However, instead of the whole TCAMS malaria box, it refers to the dataset formed by the first 10 compounds in the TCAMS Malaria Box only.
Now that we have the dataset URI, let us find the URI of a model to apply. Ideally in a new tab of your browser, go to the list of OpenTox models at http://apps.ideaconsult.net:8080/ambit2/model (or follow the “Models” link at the top of the page listing the datasets). To predict oral toxicity we will use the “Toxtree Cramer rules” model. Clicking on the Cramer rules link will open its page. Again, you can get the model URI (http://apps.ideaconsult.net:8080/ambit2/model/2) from the address bar of your browser.
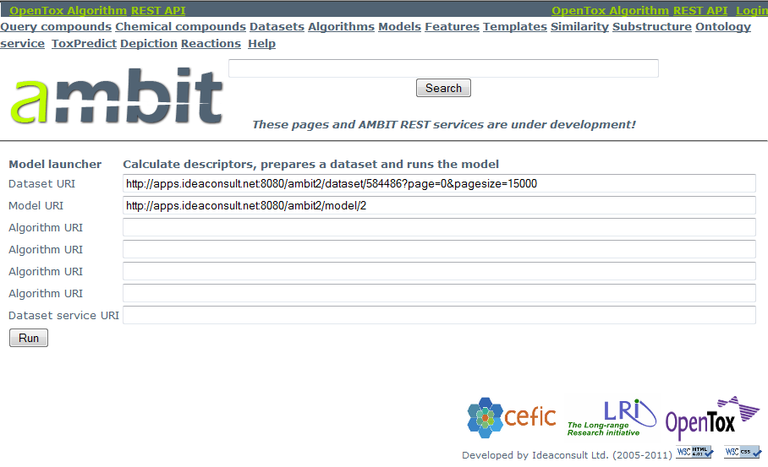
To apply the model to the TCAMS dataset there are two options: 1) you could directly paste the dataset URI into the "Dataset URI" field on the model's page, forcing the predictions to be calculated for each molecule, or 2) you could use a so-called SuperService. The preferred option is 2), because it first checks whether the model has already been applied to the dataset (in which case it simply retrieves the stored results), before actually launching calculations. To find the SuperService, navigate to the "Algorithms" page in Ambit2 (it's the fourth link from the left, at the top of the page) http://apps.ideaconsult.net:8080/ambit2/algorithm. Beneath the Ambit logo you will find a list of algorithm categories, among which you will find the link to the SuperService (http://apps.ideaconsult.net:8080/ambit2/algorithm?type=SuperService) and click on it. On the page that opens, click on the "Calls a remote service" link, which brings you to the input page for the SuperService (http://apps.ideaconsult.net:8080/ambit2/algorithm/superservice).
Enter (or paste) the TCAMS URI (“http://apps.ideaconsult.net:8080/ambit2/dataset/584486”) into the "Dataset URI" text box. IMPORTANT: Currently, AMBIT has a default upper limit of 10000 compounds when applying models to datasets. To ensure that all compounds of the TCAMS dataset are predicted, add "?max=15000" or "?page=0&pagesize=15000" to the end of the dataset URL (http://apps.ideaconsult.net:8080/ambit2/dataset/584486?max=15000 or http://apps.ideaconsult.net:8080/ambit2/dataset/584486?page=0&pagesize=15000, respectively).
Enter the Model URI (http://apps.ideaconsult.net:8080/ambit2/model/2) into the "Model URI" text box. Clicking “Run” will launch the calculations.
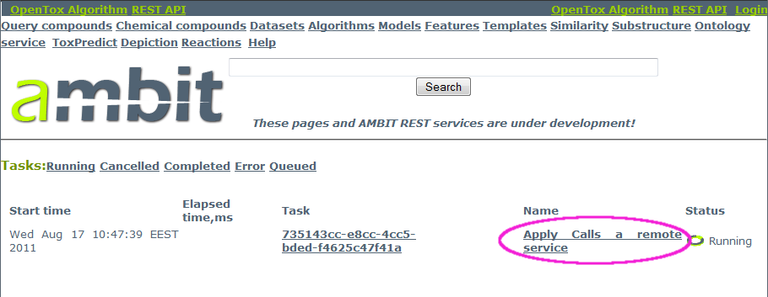
You can click on the link under "Name" (see example inside the magenta ellipse in the screenshot below) to find out if the calculations are completed. When completed, clicking on the link will lead to a dataset with the results.
The Cramer rules model is an implementation of Cramer et al., Estimation of Toxic Hazard - A Decision Tree Approach, J Cosmet Toxicol, Vol. 16, pp. 255-276, Pergamon Press, 1978. It comprises 33 structural rules and places evaluated compounds into one of three classes:
• Class I substances are simple chemical structures with efficient modes of metabolism suggesting a low order of oral toxicity;
• Class III substances are those that permit no strong initial presumption of safety, or may even suggest significant toxicity or have reactive functional groups; and finally,
• Class II are intermediate. This model is very conservative and places most of the compounds in Class III.
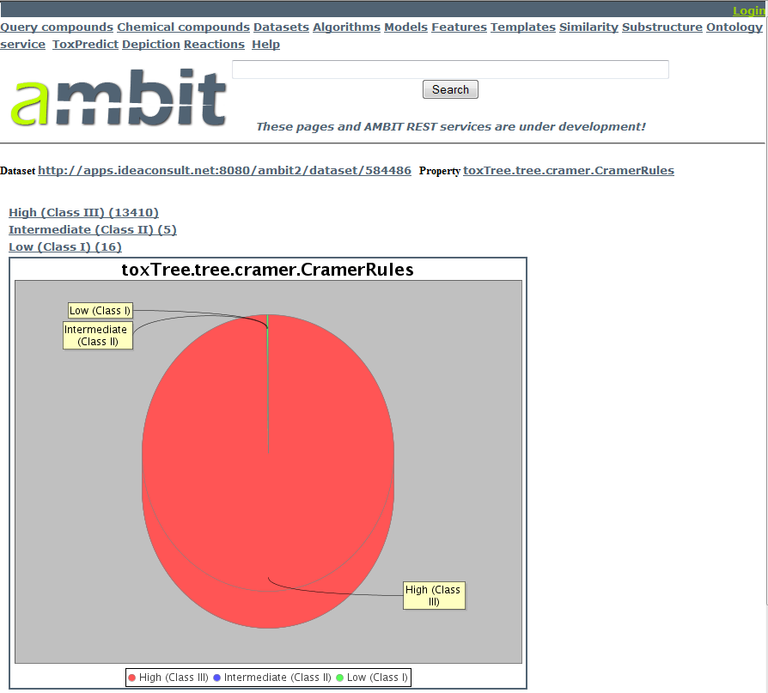
During this exercise, we’ll look for compounds of low toxicity (Class I) and high antimalarial activity. There are a small number of Class I compounds, the distribution can be seen via the OpenTox chart generation service. The chart generation service takes as input a dataset URI and a feature URI. In our case, we want to use the prediction feature URI of the Cramer rules model. To find out the feature URI of this model, navigate to the model's page in Ambit2 (http://apps.ideaconsult.net:8080/ambit2/model/2). On the model's page, click on the "Predicted" link at the very right. Following this link brings up the page with the two prediction features of the Cramer rules model: the actual prediction into one of the three classes, and an explanation of the prediction. We're interested in the class predictions. Thus, click on the "toxTree.tree.cramer.CramerRules" link at the top-left of the table. From the resulting feature page, note the feature URI in the address bar of your browser: http://apps.ideaconsult.net:8080/ambit2/feature/22254.
We have all we need to generate a chart now. The chart generation service is at http://apps.ideaconsult.net:8080/ambit2/query/ncompound_value, the dataset URI is dataset_uri=http://apps.ideaconsult.net:8080/ambit2/dataset/584486, and the feature URI is http://apps.ideaconsult.net:8080/ambit2/feature/22254. Combining these leads to:
As you can see from the chart, only very few compounds are classified into Class I by Cramer rules. To continue working with these compounds, we need to have a way to filter the dataset according to this prediction feature. As there is currently no interface for such a search, we have to construct a URI to carry out this search/filtering. The URI construct is as follows:
http://apps.ideaconsult.net:8080/ambit2/dataset/<number>?property=http://apps.ideaconsult.net:8080/ambit2/feature/<number>&search=<search query>
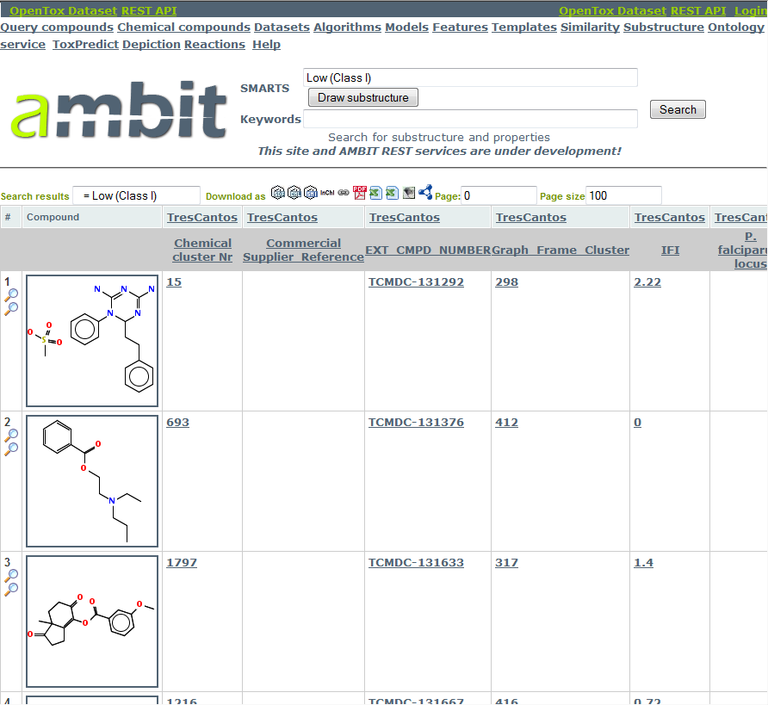
Simply replace <number> with the dataset and feature number within which you'd like to search, and the <search query> with what you want to search for. In our case, the dataset number is 584486, the feature number is 22254 (we know that from the chart generation step), and our search query is "Low (Class I)". This query is problematic for URIs because of the spaces. Replace them with "+" symbols, turning the search query into "Low+(Class+I)".
The resulting URI is:
Step 2: Analyse Cytotoxicities of the Cramer Class I compounds