Load a model and carry out a prediction
Specify a model using its OpenTox URI
In the next tab of Q-edit, the "Model" tab, the predictive model that was used to generate the prediction that is reported needs to be specified. If you know the URI of an OpenTox model resource, you may enter it directly to the corresponding "OpenTox Model URI" field in Q-edit:
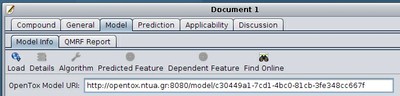
How to obtain the URI of an OpenTox model
OpenTox services list the URIs of all available models under /model. For instance check out http://apps.ideaconsult.net:8080/ambit2/model or http://opentox.ntua.gr:8080/model. Alternatively, you can search for approved models that are included in the OpenTox application ToxPredict (http://toxpredict.org) at http://apps.ideaconsult.net:8080/ontology.
To obtain the model URI e.g. from the list of models presented at http://apps.ideaconsult.net:8080/ambit2/model, simply navigate to http://apps.ideaconsult.net:8080/ambit2/model with your browser, choose a model and click on its name (in the "Model name" column). The page of the chosen model will appear. The URI of the model can now be obtained from the address bar of the browser.
Load the model description from its URI
Once an OpenTox model URI has been entered to Q-edit, click the "Load" button or strike enter to load all information available for it. In case the model you chose is password-protected you will need to provide your credentials. The following dialog box will appear:
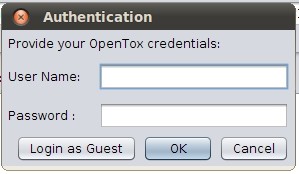
In case you don't have an OpenTox account, you may just "Login as Guest" clicking on the corresponding button.
Now you can inspect details about the loaded model - just click on the "Details" button that appears in the "Model" tab toolbar:
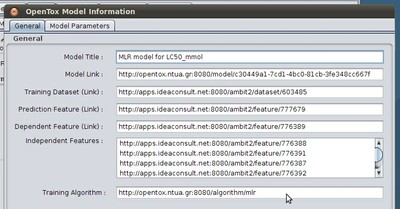
In the model details one can find the Model's title (e.g. here it is "MLR model for LC50_mmol"), the prediction features as well as the dependent and independent features of the model, or a link to its training dataset. All these details are identified by their URIs. Later we will show how to access more human-readable information about all these.
If the model has a certain parametrization, this can be found in the second tab, namely "Model parameters":
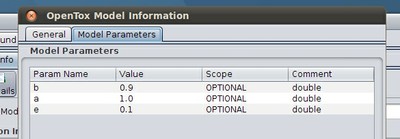
Information regarding the training algorithm that was used to create the model can be retrieved by clicking on the "Algorithm" Button, and the same way one can inspect all available information for the predicted and dependent features of the model.
Apply the model to obtain a prediction
Use the chosen model to obtain a prediction for the selected compound. Using the compound and the QSAR model provided by the user in the previous sections, Q-edit can invoke the necessary OpenTox web services to return the predicted value as well as (if available) the experimental value for the compound. This functionality is available under the fourth tab in the Q-edit main view - "Prediction" - where the user is required to report the predicted value.
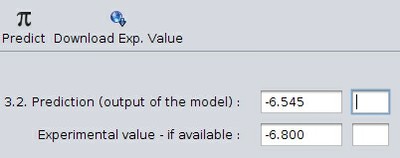
(note that the empty fields in the image above hold the units of the predicted and experimental values - if available.)
The user is also expected to comment on the uncertainty of the prediction and the possible chemical and/or biological mechanisms that might affect its reliability (for example, a particular compound might appear to be active according to a QSAR model but in practice it is not well absorbed). This information can be entered using the "Discussion" button in the toolbar of the "Predict" tab.
--
Next: Applicability domain and structural analogues - How reliable is the prediction?
Back: Provide General Information - Specify authors and date of the report

