Prioritizing Compounds, Step 3: Predicting the Mutagenicity of the Selected Compounds
Predicting the Mutagenicity of the Selected Compounds
To add a further criterion to be used when selecting our drug candidate, we predict the compounds’ mutagenicities. To do so, we’ll use the Toxtree Benigni/Bossa rules for mutagenicity and carcinogenicity (Benigni et al., Mechanistic QSAR of aromatic amines: new models for discriminating between mutagens and nonmutagens, and validation of models for carcinogens, Environ Mol. Mutag. 48:754-771 (2007).). The URL of this model is http://pirin.uni-plovdiv.bg:8080/malaria/model/12.
Analogously as you have done for the Cramer rules, follow the URL of the Benigni/Bossa model (http://pirin.uni-plovdiv.bg:8080/malaria/model/12), type or paste the URL or the TCAMS dataset (http://pirin.uni plovdiv.bg:8080/malaria/dataset/12) in the text box and click “Predict”. Alternatively, the URL of the filtered list of the previous step could be entered here, as well.
OpenTox models store the prediction results again under data columns with unique URL. These are available via http://host/model/{id}/predicted , which in our example corresponds to
http://pirin.uni plovdiv.bg:8080/malaria/model/12/predicted
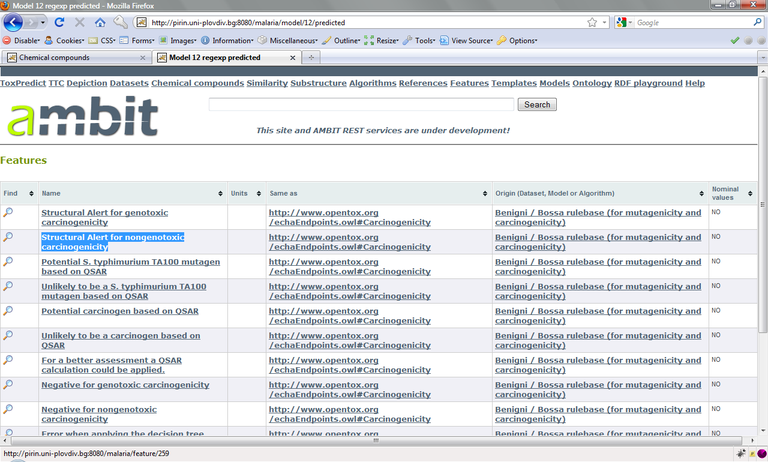
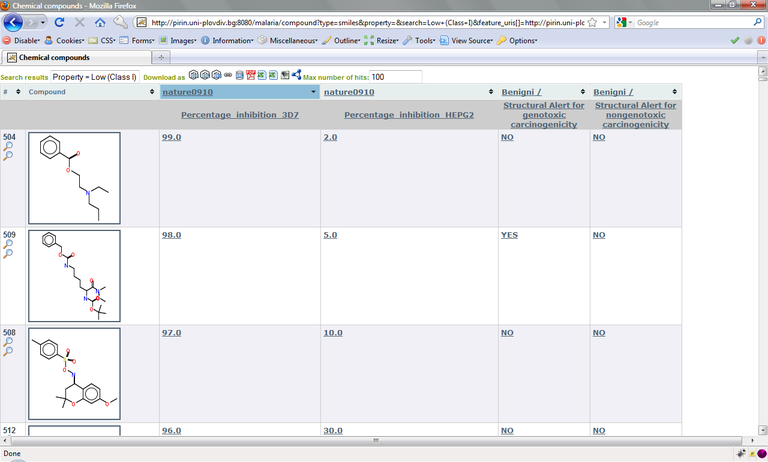
For our purpose, we select the columns “Structural Alert for genotoxic carcinogenicity“ (http://pirin.uni plovdiv.bg:8080/malaria/feature/258) and ”Structural Alert for nongenotoxic carcinogenicity“ (http://pirin.uni plovdiv.bg:8080/malaria/feature/259). As before, we add data columns for these structural alerts to our Cramer-class filtered list of compounds, again using the feature_uris[] method. The resulting URL is:
http://pirin.uni plovdiv.bg:8080/malaria/compound?type=smiles&property=&search=Low+(Class+I)&feature_uris[]=http://pirin.uni plovdiv.bg:8080/malaria/feature/190&feature_uris[]=http://pirin.uni plovdiv.bg:8080/malaria/feature/179&feature_uris[]=http://pirin.uni plovdiv.bg:8080/malaria/feature/258&feature_uris[]=http://pirin.uni plovdiv.bg:8080/malaria/feature/259
The resulting table (as well as any other) can be sorted according to the values in any column by clicking on the column header.
In the following examples, we’ll consider the first compound in the image below as our antimalarial drug candidate. It is a Cramer class I compound that inhibits growth of P. falciparum 3D7 by 99% at the concentration tested (2µM), has a very low human cytotoxicity and no structural alerts for carcinogenicity. (You may choose a different compound).
Similarly to datasets and models, each compound in OpenTox services also has its unique URL. You can find the URL of a compound by clicking on its 2D structure, and stripping off the “?media=text/html” part at the end of the URL this brings you to.
The URL of the compound selected above is http://pirin.uni plovdiv.bg:8080/malaria/compound/458166/conformer/773441.