Predict Kinase Inhibitor Activity, Step 1: Selecting a Subset to Create a Model with ToxCreate
Selecting a subset of compounds to create a model with ToxCreate
To create the dataset required for model building go to http://pirin.uni plovdiv.bg:8080/malaria/dataset
Click on “Tres Cantos Antimalarial TCAMS dataset (screening against GlaxoSmithKline's library)“ (http://pirin.uni-plovdiv.bg:8080/malaria/dataset/12)
Browse the dataset and find the column “Target hypothesis”. You will note that most entries are empty (only ~6% of the compounds have a target hypothesis annotated). In the 100 compounds displayed by default when following the link to the TCAMS data, you will only find one entry with value “Adrenergic receptor antagonist“ (http://pirin.uni-plovdiv.bg:8080/malaria/compound?feature_uris%5b%5d=http://pirin.uni-plovdiv.bg:8080/malaria/feature/183&property=Target_Hypothesis&search=Adrenergic+receptor+antagonist&feature_uris%5b%5d=http://pirin.uni-plovdiv.bg:8080/malaria/dataset/12/feature). You could click on the link, which would filter out only compounds with this potential target.
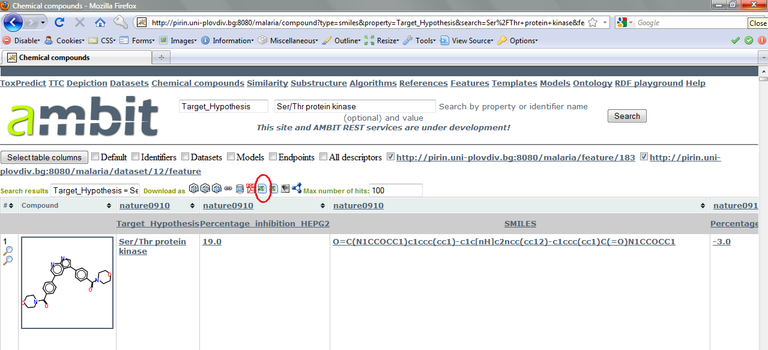
For our purpose, we want the list of compounds annotated to be kinase inhibitors. You could try to increase the number of displayed compounds until you find one, or you could enter “Ser/Thr protein kinase” in the searching text box at the top of the page and click the “Search” button. The results will be displayed as below (see Figure below).
To build a model, it is not enough to have a list of Ser/Thr kinase inhibitors. We also need some “negatives”. Although strictly speaking we don’t have any true negatives, we will use the compounds that do have a target hypothesis annotation – but one that is not “Ser/Thr kinase” – as negatives. So, we extract the whole list of compounds with non-empty target hypothesis, and replace “Ser/Thr kinase” with a “1”, and all the other target hypotheses with “0”.
To extract the list of compounds with non-empty target hypotheses, use the following URL:
http://pirin.uni plovdiv.bg:8080/malaria/compound?type=smiles&property=Target_Hypothesis&search=+&feature_uris[]=http://pirin.uni plovdiv.bg:8080/malaria/feature/183&feature_uris[]=http://pirin.uni-plovdiv.bg:8080/malaria/dataset/12/feature&max=1000&condition=!%3D
This operation is not (yet) possible via the “Search” text field (it does not allow negation, e.g. something like Target_Hypothesis !=“”), but only via the URL: briefly, the search for non-empty Target Hypothesis is done in the above URL, first with &search=+ (the “+” stands for empty) – thus searching for all the empties – and then negating the search by &condition=!%3D (%3D stands for the “=” sign, thus !%3D stands for !=, or “not equal”).
When following the above URL you’ll get a table with compounds that have a non-empty Target_Hypothesis. The next step will be to export data. Click on the left one of the two little Excel icons (when moving the mouse pointer on top of it, a small text box “text/csv” should appear) to save the selected data as CSV.
For the model building, we will use the OpenTox application ToxCreate (www.toxcreate.org). Thus, first we need to format the data as explained at www.toxcreate.org/help. That is, we leave only the SMILES column and the Target_Hypothesis column.
Now you should have the Target_Hypothesis in column 1 (or A), and the SMILES in column 2 (or B). If you are using Excel, go to the cell C2. Type
=IF(A2="Ser/Thr protein kinase"; 1; 0)
and hit “Enter”. Again click on cell C2 to activate it. Now double-click on the little black square at the bottom-right corner of the cell’s border to fill the column with this formula.
Now, copy the whole column C, and paste it (at the same place) using Excel’s “Paste Special” function, pasting only the values. Once that’s done, delete column A (holding the text entries for the Target_Hypothesis). Delete as well row 1 and save the resulting table as text CSV file to TCAMS-kinase_full.csv.
In your web browser, navigate to www.toxcreate.org. Read the instructions, and try to create a model using your dataset. As ToxCreate is currently a prototype, there are still some limitations. You might get an error in the model building, in which case you could try to reduce the number of compounds used to build the model to about 600. Just delete some rows until that table contains 600 rows or less. Save the resulting table to TCAMS-kinase-subset.csv.