Generate Predictions and their QPRF Reports
Facilitated by Pantelis Sopasakis and Sunil Chawla
Summary
The QSAR Prediction Reporting Format (QPRF) is a harmonised template for summarizing and reporting substance-specific predictions generated by (Q)SAR models (check the specification here)
The OpenTox report on Automated Validation Report Generation contains the background and progress which has been achieved within OpenTox with respect to the creation of automated reporting facilities for validated (Q)SAR models of predictive toxicology data.
In this session, we will start by reviewing the key requirements of a QPRF report and then proceed to with the following steps to generate a QPRF report using Q-edit, a new QPRF editor developed under OpenTox to facilitate the creation of QPRF reports by an end user.
Users can download the OpenTox QPRF editor (Q-edit) from http://opentox.ntua.gr/files/Q-edit.jar.
Help documentation is located at http://opentox.ntua.gr/files/Q-edit-documentation.pdf
Requirements
- Install the Java Runtime Environment on your computer.
- Download the QPRF editor (Q-edit) from http://opentox.ntua.gr/files/Q-edit.jar .
- Install the SSL certificate for opensso.in-silico.ch in your Java directory following the instructions provided at http://opentox.ntua.gr/index.php/blog/77-ssl-certificates - this is necessary to access certain protected resources of OpenTox.
Presentation Content
During the presentation we will explain how to:- Create a new (empty) QPRF report
- Authenticate oneself by providing credentials (Requires installation of the SSL certificate mentioned above).
- Search for a compound using an OpenTox web service (e.g. http://apps.ideaconsult.net:8080/ambit2 – Inspect the downloaded compound (View Chemical name(s), SMILES string, CAS RN and a depiction of the compound). View/Edit additional meta information about the compound, e.g. discuss its stereo-chemical features that might affect the validity of the prediction
- Inspect a downloaded model (Get a list of all independent features for the model); access information concerning the dependent and predicted feature; examine the training algorithm and get further (meta)information about it; find QMRF reports created for that model; add some discussion about the model
- Acquire a list of structural analogues of the compound for which the QPRF report is created and append some discussion.
- Add information about Authors of the QPRF report.
- Export the report in PDF format. The resulting document is fully compliant with the standards for QPRF reports that are provided by the EC JRC
- Save the report in binary format (as a .ro file) .
Additional material
We prompt users to download a sample PDF file of a QPRF report for the compound epicorticosterone as it was generated by Q-edit (version 1.0.12) from here. Additionally, a report file can be downloaded from here; save the file epicorticosterone.ro in a local directory and use Q-edit to open it: Launch Q-edit, Go to File > Open local resource (Ctrl + O) > Browse to the location of epicorticosterone.ro and click Open.
Notes/Remarks
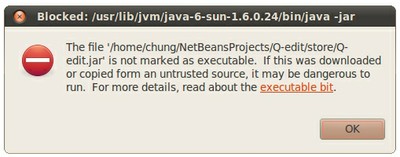
It is possible that Linux users get a warning message like the following:
You may ignore it and change the file permissions to render it executable. For this purpose you may use the command:
chmod +x ./Q-edit.jar
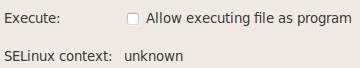
Alternatively, right click on the file Q-edit.jar, choose the Properties menu item, go to the Permissions tab and check the option "Allow executing file as program":
Complete the evaluation form following this link: Evaluation Form